Part 1
In this post I’ll be taking a look at whole genome sequencing. This is a powerful new tool for researchers and doctors. Technology today allows us to semi-cheaply sequence the entire genome of anybody. This can be used for many types of research and even by your doctor to diagnose a condition.This new technology does bring many obstacles and issues. Today I’ll be looking at one such scenario and weighing in on what would be the best plan of action for a patient with Canavan disease.
In the scenario we have a patient with Canavan disease. Some treatments have begun but they have shown a complex response to multiple medications that are not explained by the disease variant or by 1 year of traditional diagnosing. The patients family is faced with the following choices.
- Participate in a clinical trial offering full exome analysis for [Mary/John] and their parents at no personal cost.
- Seek full genome analysis and work with their insurance provider to seek coverage, a 4-6 month negotiation.
- Pay out of pocket for the full genome analysis ($5-10k).
- Use direct-to-consumer services and perform independent analysis of the raw results.
Unfortunately for Canavan disease deciding the patient’s treatment choice would most likely be a moot point. There is currently no cure for Canavan disease and no standardized treatment. Most patients only live into early childhood with a rare few living beyond that. Although this may not be true for long as there is some exciting research on the horizon in the form of gene therapy and triacetin supplementation. As for what this family should do, unfortunately I believe they will need to pick the third option. This is based purely on how sever a case of Canavan disease the patient has. There are some cases of Canavan patients living into their twenties, although this is quite rare. Their doctor could measure the patients aspartoacylase levels and get some sense of how sever the disease is. Most likely the patient would only live for about 18 months and if they have already underwent 1 year of traditional diagnoses they simply would not have time to wait for their insurance company. Also because of the severity of the disease they simply couldn’t risk the case study because it might not find the problem and they couldn’t afford wasting that time. As for the fourth option I still believe that it would simply take too much time. Unless someone in their family had experience with whole genomes and could quickly assess the issue.
Another ethical issue with whole genome analysis is incidental information. Incidental information is the information you get from a patient’s genome that they did not originally ask for. So a question is raised if this patient had his genome analysed what should the doctor do with the incidental information? I believe that this should completely be up to the patient. One argument for that sentiment is brought up in the article When Getting Your Genome is Terrifying. In the article the author speaks about how he personally would be scared to learn about major life changing health issues his genome might show. He is living a happy life and doesn’t want that to change. Personally I don’t agree with that sentiment and would learn what my genome has to say about me in a heartbeat. This would allow me to plan and hopefully prevent issues so that in the long run I could have a happier and healthier life. But I do believe that it should still be up to the patient because there are those that could not handle that information in a safe and productive way. I feel that forcing that information on a patient breaks the autonomy that patients have. While the information would most likely be beneficial a lot of people simply don’t want to have the extra burden and worry about what their genome might say about them.
Part 2
In this second part I will be writing about the variant that I have been using in previous assignments for ASPA in VCF format. This is a format for storing whole genome data that only saves the variants from the genome being analysed.
In this picture you can see the rsID given to the variant that causes Canavan disease from ASPA. The ID is rs28940279. 
In this picture you can see the rsID on the NCBI genome browser and look at its actual position on chromosome 17. The variant is at position 3,499,000 from the start of the chromosome and is located on the sixth and last exon. You can see on the second picture the exons outlined in red and the rsID labeled at the top. 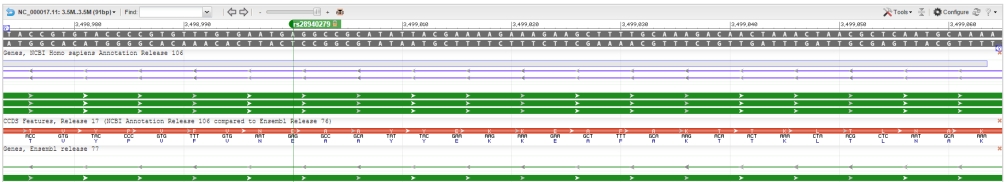
Below is the VCF format for my pet gene’s variant given the information in the assignment.
| #CHROM | POS | ID | REF | ALT | QUAL | FILTER | INFO | FORMAT | NA00001 |
| 17 | 3,499,000 | rs28940279 | A | C | 25 | PASS | NS=1; DP=35; AF=1 | GT:GQ | 0|1:52 |